Part:BBa_K1933200
constitutive expression of anti-Norovirus GII.4 scFv fused to INPNC with 6xHis tag
anti-Norovirus GII.4 scFv fused to INPNC with 6xHis tag is one of a series of surface expressing fusion proteins that make up biodevice that aims to be the therapeutic solution against norovirus infections. This protein in particular is a single chained variable fragment(scFv) of a norovirus GII.4 strain fused to surface expression anchoring domain(INPNC), connected by a 6xHis tag to be easily identified by Western blotting.
For more information, please visit [http://2016.igem.org/Team:Kyoto our wiki].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1012
Illegal XhoI site found at 1422 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 132
Illegal NgoMIV site found at 465
Illegal AgeI site found at 883 - 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
We were able to express anti-NoV antibody fragment (scFv) in the membrane fraction of E.coli using INPNC system. scFv is a protein made from fusing a variable region of the antibody, VH, and VL, with a linker peptide.
NoV binding to Histo-blood group antigen (HBGA), expressed in red blood cells and duodenum cells, is suggested to be an important part of NoV invasion. Also, NoV binding to 12A2 antibody in its HBGA binding site has been demonstrated previously. [1] The scFv coded in our part derives from antibody 12A2, so it also sterically blocks the binding of NoV to HBGA in theory, which might neutralize NoV using the same mechanism employed by human immune system. We created this parts for the realization of a biodevice to remove NoV from human intestine or from the environment.
Characterization
Sequence confirmed!
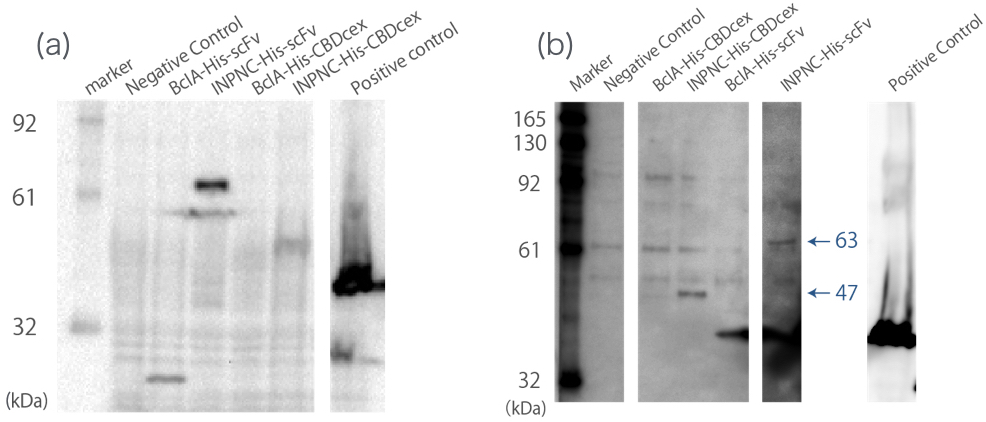
Western blotting
We used whole-cell Westernlotting with anti-His tag antibody(Fig.1(a)). Then, we prepared the membrane fraction from the E. coli lysate. Membrane fraction was solubilized and used for Nickel Sepharose purification and precipitates were examined by Western blotting against His tag(Fig.1(b)).
A band corresponding to INPNC-His-scFv (63 kDa) was observed both in whole cell and in membrane fraction Western blotting (Fig.1(a),(b)), which confirms expression of the fusion proteins, and suggests that scFv was surface expressed, respectively.
Scanning Electron Microscopy
First, we constructed INPNC-His-scFv and BclA-His-scFv (BclA:another surface display module) encoding plasmids. Then we transformed E. coli (strain:DH5α)with these plasmids, and observed its interaction with NoV-like particles (NoVLP, the capsid proteins of NoV) of NoV GII.4 strain. (Fig.2) We observed NoVLP only on the surface of INPNC-His-scFv expressing E. coli , as NoVLP could not be observed in E.coli expressing BclA-His-scFv, (Which may be due to low expression levels, seen in Fig.2), this NoVLP is not unspecifically mixed with the sample. Thus we concluded that we observed the binding of our INPNC-His-scFv expressing E. coli to NoVLP.
Thus, we conclude that this part successfully worked.
- NoVLP was kindly given to us by Professor Daisuke Sano in Hokkaido University.
aspect ratio of recombinant E.coli
When comparing SEM pictures, we noticed that E. coli using INPNC-His system seems to be more elongated compared to those with BclA-His system. To confirm this, we determined aspect ratios of all E. coli from the SEM pictures using ImageJ (see Materials and Methods for more detail LINK). By Welch’s t-test (two-sided), we found that E. coli with INPNC-His system was in fact more elongated than BclA-His system expressing E. coli. (p=0.0015<0.05).
growth curve
We have also made a growth curve of the INPNC-His- and BclA-His- system expressing E. coli. It shows that INPNC-His-scFv expressing E. coli has significant delays in growth compared to its BclA counterpart. The results from growth curve and elongated shape both suggests that expression of INPNC-His-scFv to the outer membrane caused significant stress and deformation to the host E. coli.
Judging
We fulfilled criteria listed below with this part.
- [http://2016.igem.org/Team:Kyoto/Proof Proof of concept] (Gold)
- Improve a previous part or project(Gold)
Reference
[1]Shanker, Sreejesh, et al. "Structural basis for norovirus neutralization by an HBGA blocking human IgA antibody." Proceedings of the National Academy of Sciences (2016): 201609990.
| chassis | E. coli |